Multi Objective First Run
Summary
multi-objective worked and spat out a range of solutions. The solutions are on a frontier that at first sight looks not-optimal, but may be so because of the problem formulation. Skip to Fun Pictures.
This code was run and shared using jupyter. Many thanks to this post explaining how to put a jupyter session into a jekyll blog.
The evolve_colony_multi_obj.py script that generated the data is in git tag multi-obj-0.
Jupyter Session
import evolve_colony_multi_obj as ev
reload(ev)
<module 'evolve_colony_multi_obj' from 'evolve_colony_multi_obj.py'>
The imported module is a script with a main() function that runs the evolution. The fitness evaluator outputs (size, health) for the evaluated phenotype (a colony of nodes).
info = ev.main()
/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/site-packages/transforms3d/quaternions.py:400: RuntimeWarning: invalid value encountered in divide
vector = vector / math.sqrt(np.dot(vector, vector))
gen nevals avg std min max
0 25 [ 179.69142857 100.03209854] [ 158.46217436 167.08081721] [ 2. -19.99791905] [ 439.57142857 355. ]
1 48 [ 227.76571429 16.73458574] [ 143.79577287 101.42524822] [ 2. -16.86966543] [ 447. 364.64285714]
2 46 [ 233.31428571 17.55206654] [ 138.30070167 102.03801106] [ 2. -16.86966543] [ 447. 364.64285714]
3 45 [ 235.76571429 20.21989035] [ 153.93481727 102.09670425] [ 2. -16.06184076] [ 447. 364.64285714]
4 39 [ 175.50285714 38.46083977] [ 145.44557363 120.98575388] [ 2. -16.71785968] [ 448.57142857 364.64285714]
5 46 [ 173.79428571 37.89007022] [ 137.98761897 121.15668099] [ 2. -16.71785968] [ 448.57142857 364.64285714]
6 41 [ 190.66285714 39.57204475] [ 147.25656566 121.45121315] [ 2. -16.71785968] [ 448.57142857 364.64285714]
7 46 [ 148.56 61.71167985] [ 152.06825492 134.6276333 ] [ 2. -16.71785968] [ 448.57142857 364.64285714]
8 45 [ 143.56571429 87.54939393] [ 154.78536808 156.97171347] [ 2. -16.71785968] [ 448.57142857 364.64285714]
9 47 [ 173.69714286 21.5299556 ] [ 157.46930933 77.42182592] [ 2. -16.71785968] [ 448.57142857 367.85714286]
10 46 [ 160.92571429 27.50332923] [ 159.32660395 79.07821019] [ 2. -16.71785968] [ 448.57142857 367.85714286]
n_nodes = []
health = []
for individual in info.final_pop:
N,H = individual.fitness.values
n_nodes.append(N)
health.append(H)
import matplotlib.pyplot as plt
import matplotlib.cm as cm
# order by health
import numpy as np
ordered_idx = np.flip(np.argsort(health), 0)
health = np.array(health)[ordered_idx]
n_nodes = np.array(n_nodes)[ordered_idx]
v = np.linspace(0, 1, len(n_nodes))
color = cm.viridis( v )
fig = plt.figure(figsize=(9,6))
#fig = plt.figure()
plt.scatter(n_nodes, health, c=color)
plt.xlabel("Number of Nodes")
plt.ylabel("Colony Health")
plt.show(fig)
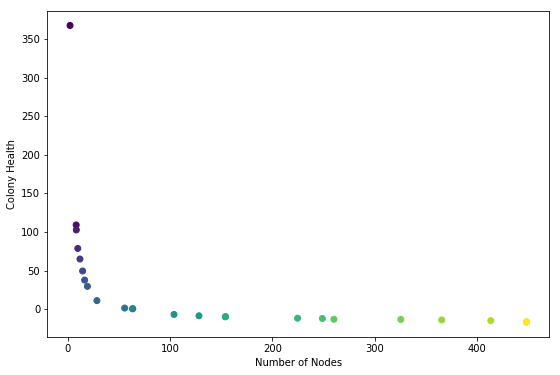
NOTE: colors are for verifying that node health ordering is correct
At first glance this was completely the reverse of what I was hoping to see. I expected to see a convex buldge pointing towards the upper right hand corner.
Hypothesis 1:
It looks as if the ea is trying to minimize the number of nodes and the colony health. This is not what the code specifies.
Hypothesis 2:
I did set up a system where high health is likely to be achived by a colony with a low number of nodes, and vice-versa. It could be that this curve, which looks like an inverse-relationship, is a result of the mechanics of the system. It might be inevitable. If that is the case I would expect to see the curve bump out generation by generation.
# Save an image for each genome in the final population
import mayavi.mlab as mlab
for i,idx in enumerate(ordered_idx):
genome = info.final_pop[idx]
p = ev.make_phenotype(genome)
p.show_lines()
mlab.savefig(str(i).zfill(2)+'_genome_'+str(idx)+'.png')
mlab.close(all=True)
health_rank = 15
idx = ordered_idx[health_rank]
genome = info.final_pop[idx]
p = ev.make_phenotype(genome)
p.number_of_elements()
151
p.get_health()
-10.443708609271523
#print(genome)
p.show()
Below is a screen shot of the 3d view generated by show():
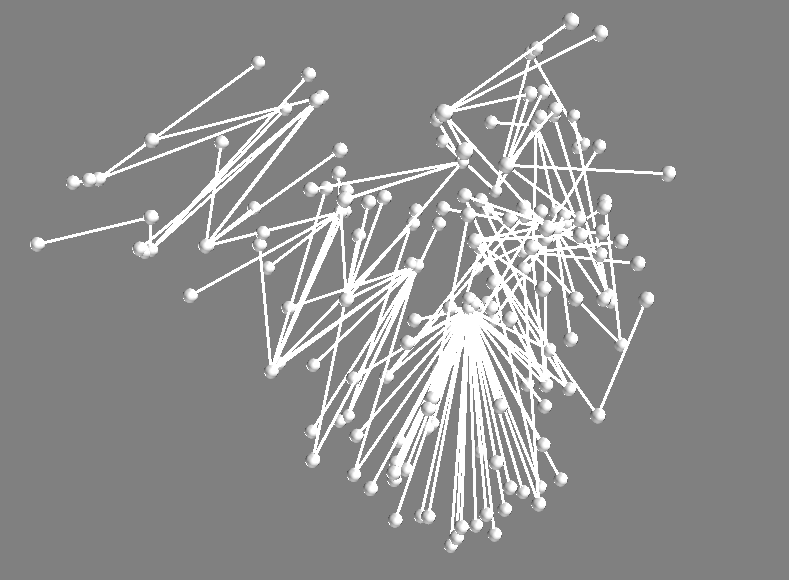 It makes sense that this has low health! The nodes below are being starved for nutrients. But it has many nodes. After inspecting alot of the solutions, I think the evolution may be working right after all (looks like Hypothesis 2 is pulling ahead). Next steps are to make a compiliation of the images for all of the solutions in this final population. Edit just made this image. See the next post.
It makes sense that this has low health! The nodes below are being starved for nutrients. But it has many nodes. After inspecting alot of the solutions, I think the evolution may be working right after all (looks like Hypothesis 2 is pulling ahead). Next steps are to make a compiliation of the images for all of the solutions in this final population. Edit just made this image. See the next post.
here is another one
from health_rank = 9:
 (number of nodes = 24, health = 19 )
(number of nodes = 24, health = 19 )